Analytic and Web Tools Development for Single Cell Data Analysis and Visualization
As any of the emerging technologies, scRNA-seq offer unique capabilities at same time brought new challenges. There is not many mature commercial software for data analysis currently available, processing large amounts of single cell RNA-seq data from heterogeneous cell populations creates the urgent need for readily accessible tools for the data analysis and user friendly web-tools for data search and visualization. With the funding support of NIH LungMAP consortium, we developed a number of well received analytic and web-tools that are freely available to the research community.
The pipeline supports the analysis for: 1) the distinction and identification of major cell types; 2) the identification of cell type specific gene signatures; and 3) the determination of driving forces of givencell types. SINCERA is implemented in R, licensed under the GNU General Public License v3, and freely available from
The algorithm utilizes single-cell RNA-seq (scRNA-seq) to quantitatively measure cellular differentiation states based on single cell entropy and predict cell differentiation lineages via the construction of entropy directed cell trajectories.
- LungGENS: A web-based tool for mapping single cell gene expression in the developing lung
- LGEA: an integrative web portal for comprehensive gene expression data analysis in lung development
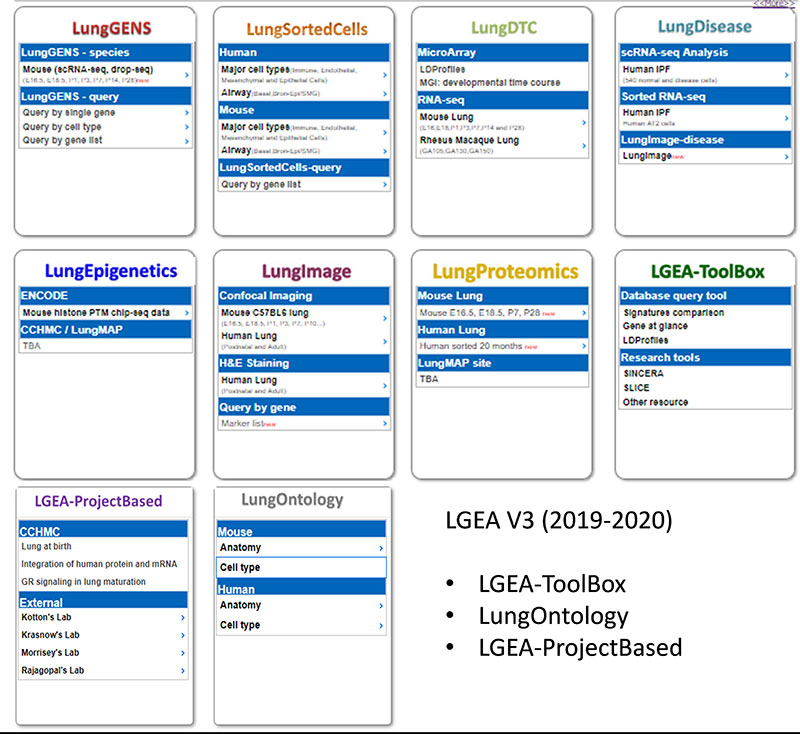
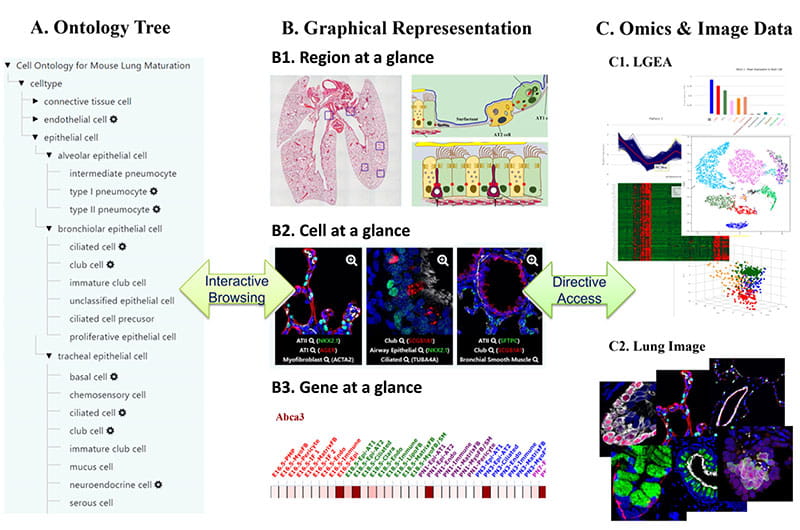
‘LungGENS’, our previously developed web tool for mapping single-cell gene expression in the developing lung, has been well received by the pulmonary research community. With continued support from the ‘LungMAP’ consortium, we extended the scope of the LungGENS database and released ‘LGEA’ to accommodate transcriptomics and other omics data from pulmonary tissues and cells from human and mouse at different stages of lung development. The current LGEA web portal provides a variety of query and analytic tools including “LungGENS”, “LungSortedCells”, “LungDTC”, “LungDiseases”, “LungEpigenetics”, “LungImage”, “LungProteomics”, “LGEA-ToolBox”, “LGEA-ProjectBased” and “LungOntology”. We are actively expending LGEA database to including more lung disease data and developing new user-friendly tools for data access, query and visualization.
Grant Support
U01 HL148856 (Whitsett/Potter/Xu)
U24 HL148865 (White, Aronow)
Chan Zuckerberg Foundation- HCA Lung Seed Network (Xu)